Reference: 1982 IUPAC Recommendations
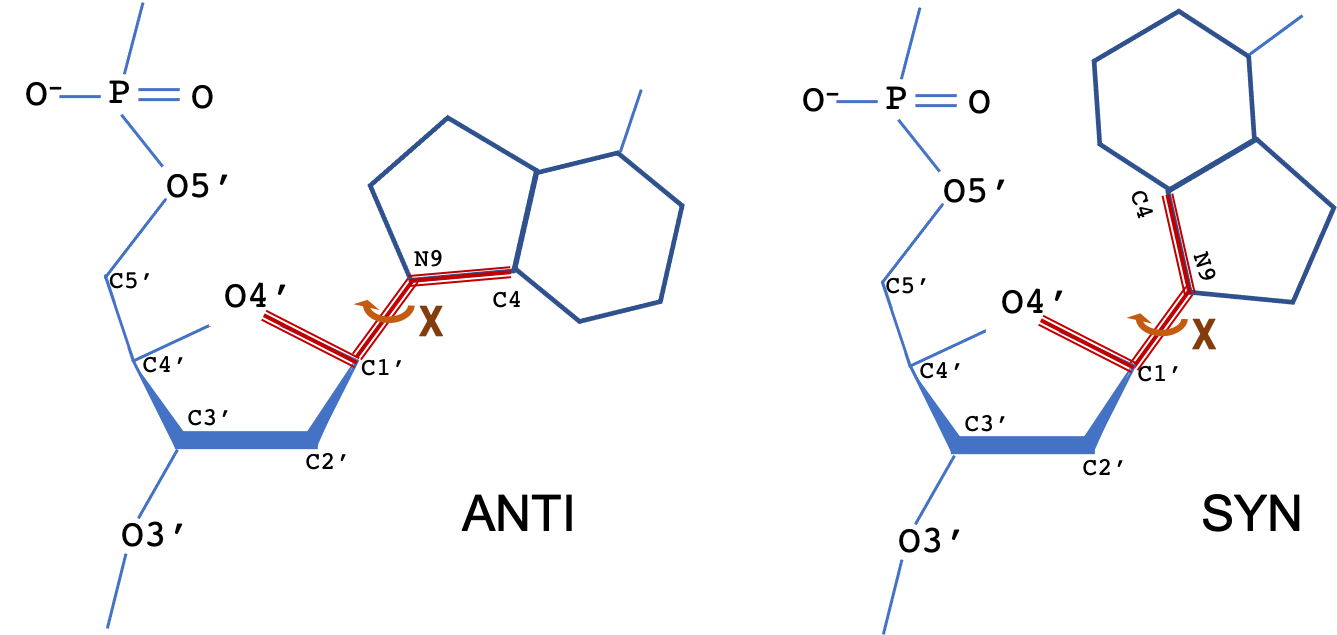
The backbone of a polynucleotide chain consists of a repeating unit of six single bonds as shown: P-O5', O5'-C5', C5'-C4', C4'-C3', C3'-O3' and O3'-P. The torsion angles about these bonds are denoted, respectively, by the symbols α (alpha), β (beta), γ (gamma), δ (delta), ε (epsilon), ζ (zeta). The χ (chi) torsion angle that describes the N-glycosidic bond is also shown.
The sugar ring occupies a pivotal position in the nucleotide unit because it is part of both the backbone and the side chain. The sugar ring is usually non-planar, and is characterized by its pucker conformation. The most common pucker conformation for the ribose sugar in RNA is C3'-endo; the most common pucker conformation for the deoxyribose sugar in DNA is C2'-endo. Other conformations are possible, see IUPAC recommendations (link above) for further information. Sugar torsions v0-v4, amplitude, and pseudorotation phase angle parameters are as defined by Altona & Sundralingham (1972). Dperpendicular (Chen et al 2010) and ssZp (Li et al 2019) are two alternate proposed geometrical methods to assess sugar pucker.
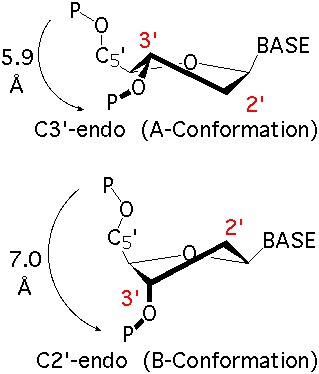
Image Source: X3DNA-DSSR Website
| Torsion | Definition |
|---|---|
| v0 | C4'-O4'-C1'-C2' |
| v1 | O4'-C1'-C2'-C3' |
| v2 | C1'-C2'-C3'-C4' |
| v3 | C2'-C3'-C4'-O4' |
| v4 | C3'-C4'-O4'-C1' |
The torsion angle about the N-glycosidic bond (N-C1') that links the base to the sugar is denoted by the symbol χ. The terms syn and anti describe different conformational regions of χ: syn (0±90°), anti(180±90°). For pyrimidine bases, χ is defined as the torsion angle for atoms O4′-C1′-N1-C2; for purine bases (as shown), the torsion angle is for atoms O4′-C1′-N9-C4.
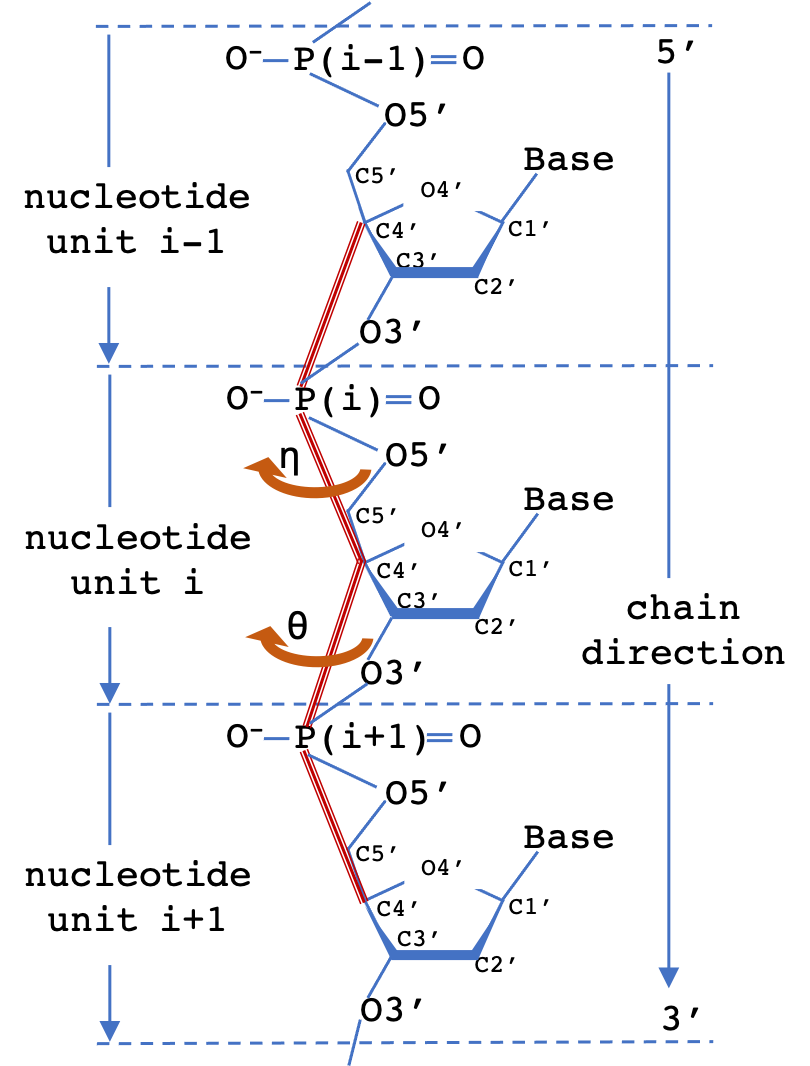
A common scheme to simplify the representation of nucleotide polymer backbone geometry is to define two virtual torsion angles (pseudotorsions), eta (η) and theta (θ) for a nucleotide triplet. The measured (η,θ) pair defines the backbone conformation relative to central residue (i). 2D Virtual torsion plots for structures containing RNA can be obtained from the EMDataResource pseudo-torsion plot server.
DNA and RNA nucleotide polymer conformation can be analysed by classification of geometry at the level of the dinucleotide step. The classification system used by Cerny et al (2016, 2020) is an extension of an earlier analysis of RNA conformation by Richardson et al (2008). The following are measured for cluster analysis: (i) seven backbone torsions: δ1, ε1, ζ1, α2, β2, γ2, δ2, (ii) two torsions around the glycosidic bonds: χ1, χ2, (iii) one pseudo-torsion angle μ involving atoms defining the χ1 and χ2 glycosidic bonds, (iv) two pseudo-distances: N1-N2 and C1'-C2'. The 96 defined dinucleotide conformational classes describe a wide range of steps with bases that are stacked or unstacked, or belonging to intercalated or open conformations. Further information and conformation analysis tools are available at the DNATCO website.
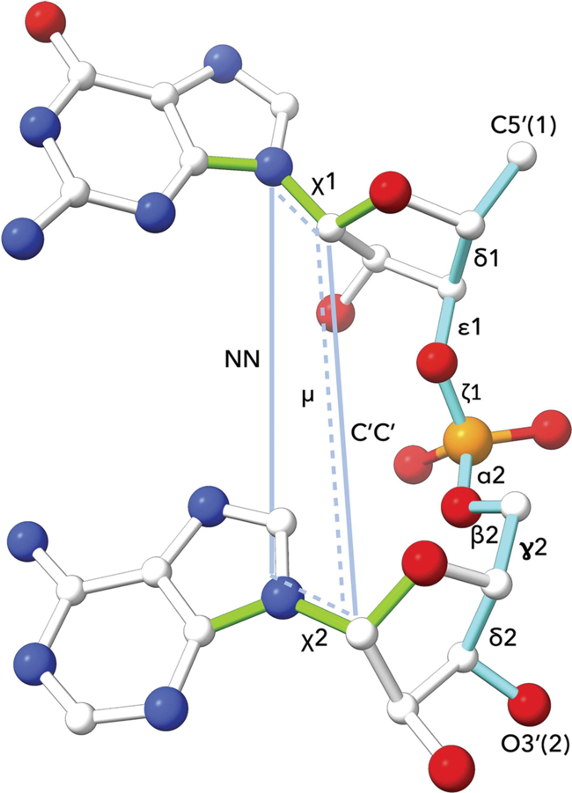
Image source: Cerny et al (2020).