The Leontis-Westhof classification system annotates basepairs according to the interacting edge used on each base (Watson-Crick, Hoogsteen, Sugar), and bond orientation (Cis, Trans). The classification table is shown below, along with diagrams explaining the edge pairings and bond orientations (Figures A, B). See also: RNA Basepair Catalog
| No. | Bond Orientation | Interacting Edges (i, j)* | Symbol | Strand Orientation | Notation* |
|---|---|---|---|---|---|
| 1 | Cis | Watson-Crick/Watson-Crick | Anti-Parallel | cWW | |
| 2 | Trans | Watson-Crick/Watson-Crick | Parallel | tWW | |
| 3 | Cis | Watson-Crick/Hoogsteen | Parallel | cWH (or cHW) | |
| 4 | Trans | Watson-Crick/Hoogsteen | Anti-Parallel | tWH (or tHW) | |
| 5 | Cis | Watson-Crick/Sugar | Anti-Parallel | cWS (or cSW) | |
| 6 | Trans | Watson-Crick/Sugar | Parallel | tWS (or tSW) | |
| 7 | Cis | Hoogsteen/Hoogsteen | Anti-Parallel | cHH | |
| 8 | Trans | Hoogsteen/Hoogsteen | Parallel | tHH | |
| 9 | Cis | Hoogsteen/Sugar | Parallel | cHS (or cSH) | |
| 10 | Trans | Hoogsteen/Sugar | Anti-Parallel | tHS (or tSH) | |
| 11 | Cis | Sugar/Sugar | Anti-Parallel | cSS | |
| 12 | Trans | Sugar/Sugar | Parallel | tSS |
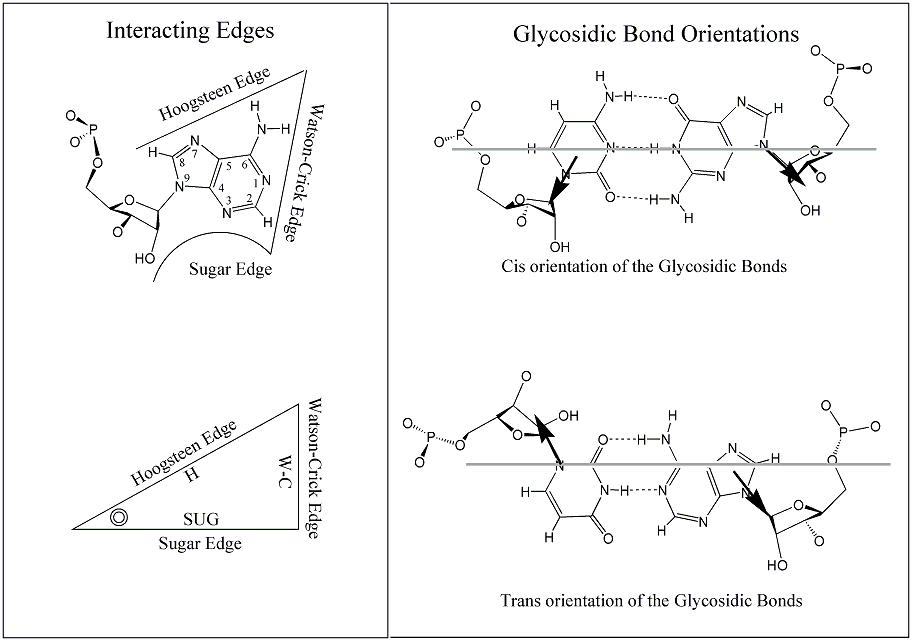
Figure A: Base edges and Base-pair geometric isomerism. (Upper left) An adenosine showing the three base edges that are available for hydrogen-bonding interactions: Watson-Crick (W-C), Hoogsteen and Sugar-edge. (Lower left) Representation of RNA base as a triangle. The position of the ribose is indicated with a circle in the corner defined by the Hoogsteen and Sugar edge. (Right) Cis and Trans base-pairing geometries, illustrated for two bases interacting with W-C edges. (Leontis & Westhof, 2001).
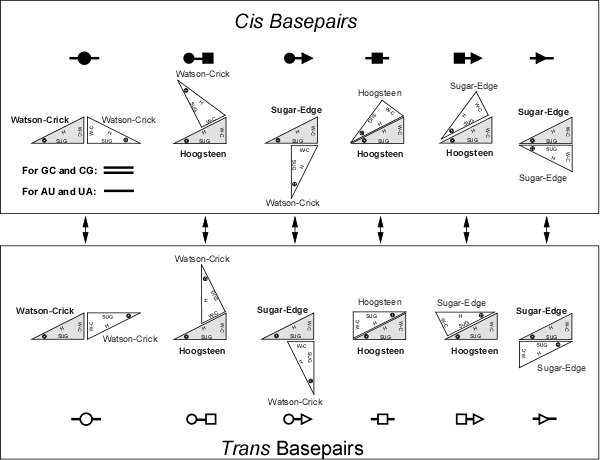
Figure B: Basepairs geometric families and their annotation. Upper panel: Twelve geometric basepair families resulting from all combinations of edge-to-edge interactions of two bases with cis or trans orientation of the glycosidic bonds. Circles represent W-C edges, squares Hoogsteen edges, and triangles Sugar edges. Basepair symbols are composed by combining edge symbols, with solid symbols indicating cis basepairs and open symbol, trans basepairs. Lower Left: Symbols for other pairwise interactions (Leontis et al., 2002).
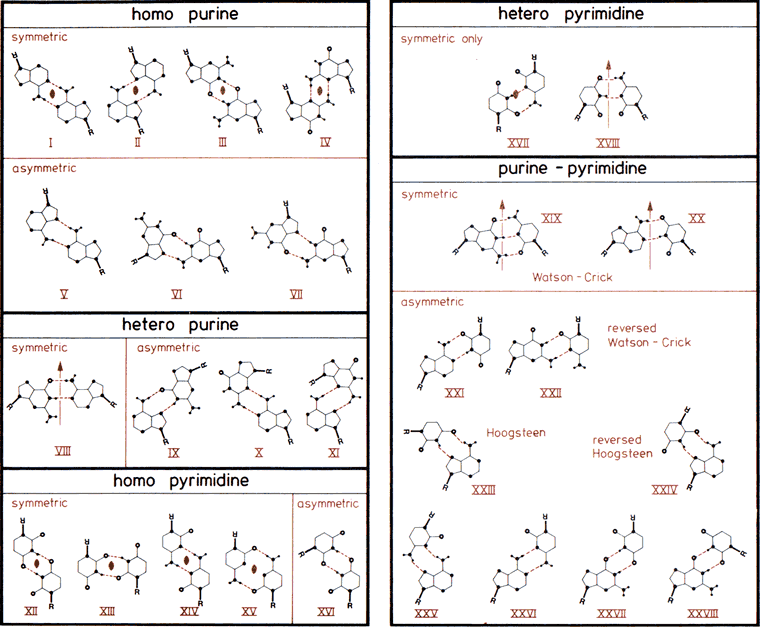
The 28 possible base-pairs for A, G, U(T), and C involving at least two (cyclic) hydrogen bonds. Hydrogen and nitrogen atoms displayed as small and large filled circles, oxygen atoms as open circles, and glycosyl bonds and thick lines with R indicating ribose C1' atom. Base-pairs are boxed according to composition and symmetry, consisting of only purine, only pyrimidine, or mixed purine/pyrimidine pairs and asymmetric or symmetric base-pairs. Symmetry elements  and
and  are twofold rotation axes vertical to and within the plane of the paper. In the Watson-Crick base-pairs XIX and XX and in base-pairs VIII and XVIII, pseudosymmetry relating only glycosyl links but not individual base atoms is observed. Drawn after compilations in (33,457).
are twofold rotation axes vertical to and within the plane of the paper. In the Watson-Crick base-pairs XIX and XX and in base-pairs VIII and XVIII, pseudosymmetry relating only glycosyl links but not individual base atoms is observed. Drawn after compilations in (33,457).
The standard reference frame described in the tables and references below is commonly used for nucleic acid conformational analysis, to describe the characteristics of base-pairs, base-pair steps, and base-pair geometry relative to the local helical axis. In the schematic diagrams within the tables, the shaded edge facing the viewer denotes the minor-groove side of a base or base pair.
Images source: X3DNA-DSSR Website