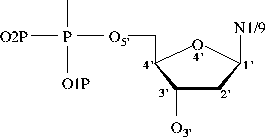
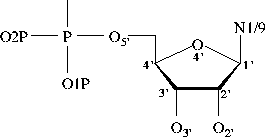
Bond Lengths in Furanose Rings
-----------------------------------------------------
ribose deoxyribose
-----------------------------------------------------
bond mean esd N mean esd N
value value
-----------------------------------------------------
C1'-C2' 1.528 (0.010, 80) 1.521 (0.014, 47)
C2'-C3' 1.525 (0.011, 80) 1.518 (0.010, 47)
C3'-C4' 1.524 (0.011, 80) 1.528 (0.010, 47)
C4'-O4' 1.453 (0.012, 80) 1.446 (0.011, 47)
O4'-C1' 1.414 (0.012, 80) 1.420 (0.013, 47)
C3'-O3' 1.423 (0.014, 80) 1.431 (0.013, 47)
C5'-C4' 1.510 (0.013, 80) 1.511 (0.008, 47)
C2'-O2' 1.413 (0.013, 80) na
C1'-N1/N9 1.471 (0.017, 80) 1.474 (0.020, 47)
(H)O5'-C5' 1.423 (0.014, 61) 1.420 (0.021, 37)
-----------------------------------------------------
-----------------------------------------------------------------------------------------------------
C2'-endo ribose C2'-endo deoxyribose C3'-endo ribose C3'-endo deoxyribose
-----------------------------------------------------------------------------------------------------
bond mean esd N mean esd N mean esd N mean esd N
value value value value
-----------------------------------------------------------------------------------------------------
C1'-C2' 1.526 (0.008, 49) 1.518 (0.010, 29) 1.529 (0.011, 24) 1.519 (0.010, 7)
C2'-C3' 1.525 (0.011, 49) 1.516 (0.008, 29) 1.523 (0.011, 24) 1.518 (0.012, 7)
C3'-C4' 1.527 (0.011, 49) 1.529 (0.010, 29) 1.521 (0.010, 24) 1.521 (0.010, 7)
C4'-O4' 1.454 (0.010, 49) 1.446 (0.010, 29) 1.451 (0.013, 24) 1.449 (0.009, 7)
O4'-C1' 1.415 (0.012, 49) 1.420 (0.011, 29) 1.412 (0.013, 24) 1.418 (0.012, 7)
C3'-O3' 1.427 (0.012, 49) 1.435 (0.013, 29) 1.417 (0.014, 24) 1.419 (0.006, 7)
C5'-C4' 1.509 (0.012, 49) 1.512 (0.007, 29) 1.508 (0.007, 24) 1.509 (0.011, 7)
C2'-O2' 1.412 (0.013, 49) na 1.420 (0.010, 24) na
C1'-N1/N9 1.464 (0.014, 49) 1.468 (0.014, 29) 1.483 (0.015, 24) 1.488 (0.013, 7)
(H)O5'-C5' 1.424 (0.016, 31) 1.418 (0.025, 25) 1.420 (0.009, 20) 1.423 (0.011, 7)
-----------------------------------------------------------------------------------------------------
Bond angles in Furanose Rings
-----------------------------------------------------
ribose deoxyribose
-----------------------------------------------------
angle mean esd N mean esd N
value value
-----------------------------------------------------
C1'-C2'-C3' 101.5 (0.9, 80) 102.7 (1.4, 47)
C2'-C3'-C4' 102.7 (1.0, 80) 103.2 (1.0, 47)
C3'-C4'-O4' 105.5 (1.4, 80) 105.6 (1.0, 47)
C4'-O4'-C1' 109.6 (0.9, 80) 109.7 (1.4, 47)
O4'-C1'-C2' 106.4 (1.4, 80) 106.1 (1.0, 47)
C1'-C2'-O2' 110.6 (3.0, 80) na
C3'-C2'-O2' 113.3 (2.9, 80) na
C2'-C3'-O3' 111.0 (2.8, 80) 110.6 (2.7, 47)
C4'-C3'-O3' 110.6 (2.6, 80) 110.3 (2.2, 47)
C5'-C4'-C3' 115.5 (1.5, 80) 114.7 (1.5, 47)
C5'-C4'-O4' 109.2 (1.4, 80) 109.4 (1.6, 47)
O4'-C1'-N1/N9 108.2 (1.0, 80) 107.8 (0.8, 47)
C2'-C1'-N1/N9 113.4 (1.6, 80) 114.2 (1.6, 47)
(H)O5'-C5'-C4' 111.6 (1.7, 61) 110.9 (2.0, 37)
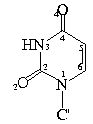
C1'-N9-C4 127.1 (1.8, 46) 125.9 (1.4, 13)
C1'-N1-C2 117.9 (1.3, 34) 117.8 (1.4, 34)
--------------------------------------------------------
-----------------------------------------------------------------------------------------------------
C2'-endo ribose C2'-endo deoxyribose C3'-endo ribose C3'-endo deoxyribose
-----------------------------------------------------------------------------------------------------
angle mean esd N mean esd N mean esd N mean esd N
value value value value
-----------------------------------------------------------------------------------------------------
C1'-C2'-C3' 101.5 (0.8, 49) 102.5 (1.2, 29) 101.3 (0.7, 24) 102.4 (0.8, 7)
C2'-C3'-C4' 102.6 (1.0, 49) 103.1 (0.9, 29) 102.6 (1.0, 24) 102.2 (0.7, 7)
C3'-C4'-O4' 106.1 (0.8, 49) 106.0 (0.6, 29) 104.0 (1.0, 24) 104.5 (0.4, 7)
C4'-O4'-C1' 109.7 (0.7, 49) 110.1 (1.0, 29) 109.9 (0.8, 24) 110.3 (0.7, 7)
O4'-C1'-C2' 105.8 (1.0, 49) 105.9 (0.8, 29) 107.6 (0.9, 24) 106.8 (0.5, 7)
C1'-C2'-O2' 111.8 (2.6, 49) na 108.4 (2.4, 24) na
C3'-C2'-O2' 114.6 (2.2, 49) na 110.7 (2.1, 24) na
C2'-C3'-O3' 109.5 (2.2, 49) 109.4 (2.5, 29) 113.7 (1.6, 24) 112.6 (3.3, 7)
C4'-C3'-O3' 109.4 (2.1, 49) 109.7 (2.5, 29) 113.0 (2.0, 24) 112.3 (2.0, 7)
C5'-C4'-C3' 115.2 (1.4, 49) 114.1 (1.8, 29) 116.0 (1.6, 24) 115.7 (1.2, 7)
C5'-C4'-O4' 109.1 (1.2, 49) 109.3 (1.9, 29) 109.8 (0.9, 24) 109.8 (1.1, 7)
O4'-C1'-N1/N9 108.2 (0.8, 49) 108.0 (0.7, 29) 108.5 (0.7, 24) 108.3 (0.3, 7)
C2'-C1'-N1/N9 114.0 (1.3, 49) 114.3 (1.4, 29) 112.0 (1.1, 24) 112.6 (1.9, 7)
(H)O5'-C5'-C4' 111.7 (1.9, 61) 110.9 (1.7, 25) 111.5 (1.6, 20) 111.0 (2.5, 7)
C1'-N9-C4 127.4 (1.2, 28) 126.3 (1.2, 7) 126.3 (2.8, 12) 123.9 (1.0, 2)
C1'-N1-C2 118.5 (1.1, 21) 117.8 (1.8, 22) 116.7 (0.6, 12) 117.5 (1.4, 5)
----------------------------------------------------------------------------------------------------
Torsion Angles in Furanose Rings
-----------------------------------------------------------------------------------------------------------
C2'-endo ribose C2'-endo deoxyribose C3'-endo ribose C3'-endo deoxyribose
-----------------------------------------------------------------------------------------------------------
torsion angle mean esd N mean esd N mean esd N mean esd N
value value value value
-----------------------------------------------------------------------------------------------------------
C4'-O4'-C1'-C2' 339.2 (5.2, 49) 340.7 (6.3, 27) 2.8 (6.1, 24) 6.6 (3.1, 5)
O4'-C1'-C2'-C3' 35.2 (3.4, 49) 32.8 (4.9, 27) 335.4 (4.9, 24) 333.3 (2.1, 5)
C1'-C2'-C3'-C4' 324.6 (2.8, 49) 326.9 (3.6, 27) 35.9 (2.8, 24) 35.6 (1.3, 5)
C2'-C3'-C4'-O4' 24.2 (4.4, 49) 22.6 (4.5, 27) 324.7 (3.0, 24) 327.7 (2.3, 5)
C3'-C4'-O4'-C1' 357.7 (5.7, 49) 357.7 (6.1, 27) 20.4 (5.1, 24) 16.4 (3.2, 5)
C5'-C4'-C3'-C2' 263.4 (4.1, 49) 262.0 (4.1, 27) 204.0 (3.1, 24) 207.0 (2.2, 5)
C5'-C4'-C3'-O3' 147.3 (4.9, 49) 145.2 (4.0, 27) 81.0 (4.4, 24) 84.8 (4.5, 5)
O4'-C4'-C3'-O3' 268.1 (5.3, 49) 265.8 (4.3, 27) 201.8 (4.2, 24) 205.4 (4.8, 5)
C4'-O4'-C1'-N1/9 216.6 (5.4, 49) 217.7 (6.7, 27) 241.4 (6.5, 24) 245.6 (2.5, 5)
O3'-C3'-C2'-O2' 319.7 (4.2, 49) na 44.3 (4.5, 24) na
----------------------------------------------------------------------------------------------------------
C2'-endo ribose/deoxyribose C3'-endo ribose/deoxyribose
----------------------------------------------------------------------------------------------------------
mean esd N mean esd N
value value
----------------------------------------------------------------------------------------------------------
O4'-C1'-N1-C2 (anti) 229.8 (18.4, 38) 195.7 (6.6, 16)
O4'-C1'-N1-C2 (syn) 63.4 (10.2, 2) na
O4'-C1'-N9-C4 (anti) 237.0 (24.3, 18) 193.3(14.0, 12)
O4'-C1'-N9-C4 (syn) 58.6 (12.0, 15) 35.6 (0.0, 2)
----------------------------------------------------------------------------------------------------------
mean esd N
value
----------------------------------------------------------------------------------------------------------
O5'-C5'-C4'-C3 (-sc) 292.9(12.3, 12)
O5'-C5'-C4'-C3 (ap) 179.4 (6.4, 22)
O5'-C5'-C4'-C3 (+sc) 52.5 (5.7, 93)
----------------------------------------------------------------------------------------------------------
Bond Lengths and Angles in the Phosphodiester Linkage in Dinucleoside Monophosphates and Trinucleoside Diphosphates
------------------------------------------
bond mean esd N
value
------------------------------------------
P-O1P/O2P 1.485 (0.017, 30)
P-O3' 1.607 (0.012, 13)
P-O5' 1.593 (0.010, 15)
(P)O5'-C5' 1.440 (0.016, 14)
(P)O3'-C3' 1.433 (0.019, 13)
------------------------------------------
------------------------------------------
angle mean esd N
value
------------------------------------------
O1P-P-O2P 119.6 (1.5, 15)
O5'-P-O1P/O2P large* 110.7 (1.2, 16)
O5'-P-O1P/O2P small** 105.7 (0.9, 16)
O3'-P-O5' 104.0 (1.9, 13)
P-O5'-C5' 120.9 (1.6, 15)
C3'-O3'-P 119.7 (1.2, 13)
O3'-P-O1P/O2P large* 110.5 (1.1, 13)
O3'-P-O1P/O2P small** 105.2 (2.2, 13)
(P)O5'-C5'-C4' 109.4 (0.8, 14)
(P)O3'-C3'-C4' 109.3 (1.5, 13)
-----------------------------------------
*Geometries for the subset with large Ox-P-O1P/O2P angles,
where x = 3' or 5'.
**Geometries for the subset with small Ox-P-O1P/O2P angles,
where x = 3' or5'.
Torsion Angles in the Phosphodiester Linkage in Dinucleoside Monophosphates and Trinucleoside Diphosphates
-----------------------------------------
torsion angle mean esd N
value
-----------------------------------------
O3'-P-O5'-C5' (-sc) 285.3 (9.8, 8)
O3'-P-O5'-C5' (+sc) 81.0 (12.1, 5)
P-O5'-C5'-C4' 183.5 (13.0, 15)
P-O5'-C5'-C4'* 163.8 (23.0, 24)
C4'-C3'-O3'-P 214.0 (8.6, 13)
C3'-O3'-P-O5' (-sc) 289.2 (4.8, 6)
C3'-O3'-P-O5' (ap) 163.1 (0.6, 2)
C3'-O3'-P-O5' (+sc) 80.7 (14.3, 5)
-----------------------------------------
*Parameter found for mononucleotides.